
Looking for MDM2 in
the diagnostic workup
of DDLPS
DDLPS=dedifferentiated liposarcoma;
MDM2=mouse double minute 2.
What does DDLPS reveal about excess MDM2?
Diagnostic workup
Histopathology
Due to their low prevalence and heterogeneity, sarcomas present challenges for accurate diagnosis1,2
Tumor biopsy helps to determine histological type and subtype, as well as the extent of tumor necrosis3
About 90% of DDLPS are high-grade sarcomas and can present microscopically as non-distinct spindle cells and pleomorphic patterns4
In a diagnosis, differentiating DDLPS from WDLPS is important1,2
Biomarker testing
Excess MDM2 is a prominent characteristic with >90% amplification rates in DDLPS5
IHC/FISH for MDM2 testing is suggested for the diagnostic workup of any sarcoma6
FISH is more sensitive than IHC6
NGS is also an option in patients to conduct broader molecular profiling7
DDLPS is associated with a delayed diagnosis8
DDLPS is frequently seen in the retroperitoneum6
Other locations include the extremities, head or neck, pelvis, and thorax or trunk6,8
The detection of retroperitoneal tumors can be delayed due to the lack of symptoms or non-specific symptoms. Therefore, the tumor can reach a large size (>10 cm) by the time of diagnosis8
Patients with DDLPS have poor prognosis and overall survival (OS) is lowest for retroperitoneal tumors compared to other sites2,9*:
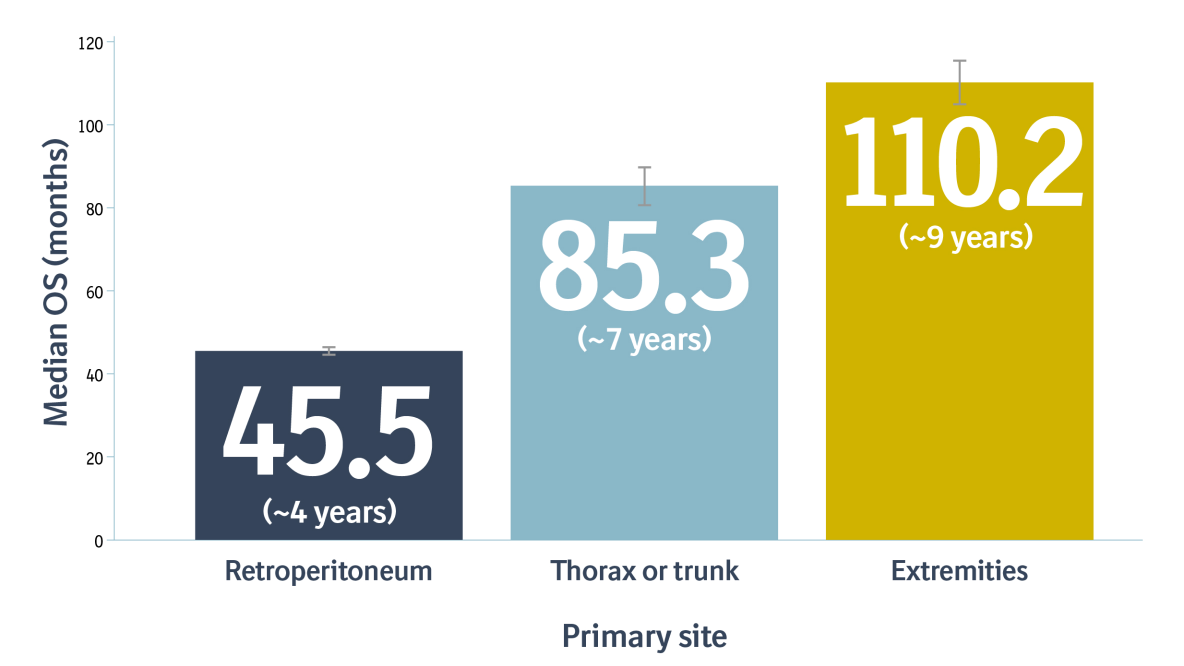
*Results from an analysis of 3024 patients with DDLPS.9
FISH=fluorescence in situ hybridization; IHC=immunohistochemistry; NGS=next-generation sequencing; WDLPS=well-differentiated liposarcoma.
References:
-
Gounder MM, Agaram NP, Trabucco SE, et al. Nat Commun. 2022;13(1):3406. doi:10.1038/s41467-022-30496-0
-
Gahvari Z, Parkes A. Curr Treat Options Oncol. 2020;21(2):15. doi:10.1007/s11864-020-0705-7
-
Gamboa AC, Gronchi A, Cardona K. CA Cancer J Clin. 2020;70(3):200-229.
-
Mariño-Enríquez A, Hornick JL, Dal Cin P, Cibas ES, Qian X. Cancer Cytopathol. 2014;122(2):128-137.
-
McGovern Y, Zhou CD, Jones RL. Front Oncol. 2017;7:292. doi:10.3389/fonc.2017.00292
-
Sciot R. Diagnostics. 2021;11(3):496. doi:10.3390/diagnostics11030496
-
Referenced with permission from the NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®) for Soft Tissue Sarcoma V.2.2023. © National Comprehensive Cancer Network, Inc. 2023. All rights reserved. Accessed August 25, 2023. To view the most recent and complete version of the guidelines, go online to NCCN.org. NCCN makes no warranties of any kind whatsoever regarding their content, use or application and disclaims any responsibility for their application or use in any way.
-
Nguyen K, Gootee J, Aurit S, Albagoush S, Curtin C, Silberstein P. Surg Case Rep. 2021;4(2):7-7. doi:10.31487/j.SCR.2021.02.02
-
Gootee J, Aurit S, Curtin C, Silberstein P. J Cancer Res Clin Oncol. 2019;145(1):181-192.